Led by Tianjin University professor Ying-Jin Yuan, a synthetic biology team at TJU has reported using the SCRaMbLE system to reveal complex rearrangements in synthetic ring chromosomes with the details of the design concept and process published in Nature Communications (DOI: 10.1038/s41467-018-06216-y) on September 17, 2018.
Genome structural variation plays a critical functional role on biological phenotypic diversity; it refers to heritable nucleotide sequence difference, and has long been interpreted with respect to human disease. Circular “ring” chromosomes have been reported in a wide variety of human genetic disorders, including epilepsy, intellectual delay, various dysmorphic features, leukemia, and microcephaly. Given the complexity of inheritance and pleiotropy associated with human ring chromosomes, model systems for the functional impact of chromosome circularization and rearrangement by design are needed. Thus, Juan Wang and Ze-Xiong Xie used the SCRaMbLE system on a synthetic ring chromosome V to study the complex genomic rearrangement of ring chromosomes, as well as the functional impacts on the phenotype.
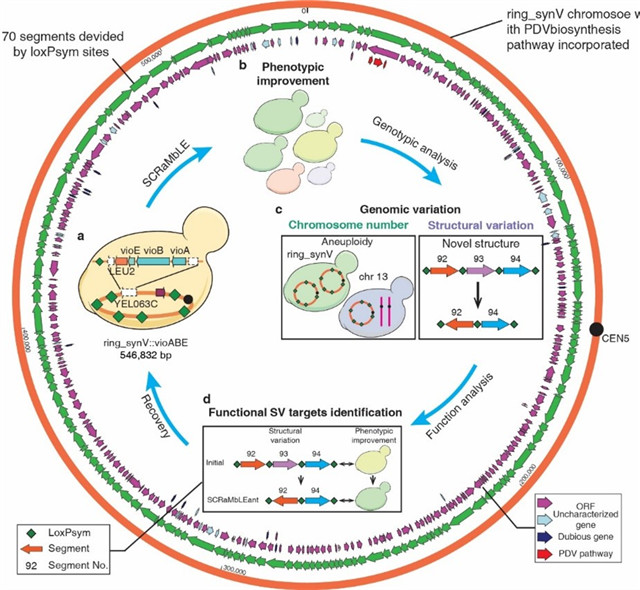
Figure. 1 Ring_synV SCRaMbLE to generate genomic and phenotypic variation
“To continuously evolve strains of defined phenotype and genotypic structural variations, we perform the inducible chromosome rearrangement by SCRaMbLEing yeast haploid cells carrying ring_synV”, said Ze-Xiong Xie, who selected PDV (a precursor of potential anti-cancer drug violacein and a downstream metabolite of aromatic amino acids) biosynthesis pathway as a selective marker to define the genotypic diversity by combinatorial rearrangements. “We isolate derivatives with a broad variety of phenotypes, facilitated by structural variants especially the large DNA segment duplication, translocation and inversion. The SCRaMbLE of a ring chromosome led to the generation of aneuploid chromosome I, III, VI, XII, XIII, and ring_synV. The size of a SCRaMbLEd ring_synV chromosome after the fifth cycle of SCRaMbLE is 101.02% larger than the initial ring_synV. We specially figured out 53 novel structural junctions. There are 29 novel junctions in the SCRaMbLEd ring_synV after the first cycle of SCRaMbLE and increased to 47 novel junctions in the SCRaMbLEd fifth cycle of SCRaMbLE, which indicate more complex structures were subsequent from the neochromsome during the continuous SCRaMbLE. We also identify and investigate if 11 SVs may be correlated with PDV metabolism improvement, identifying targets for phenotypic comprehension.”
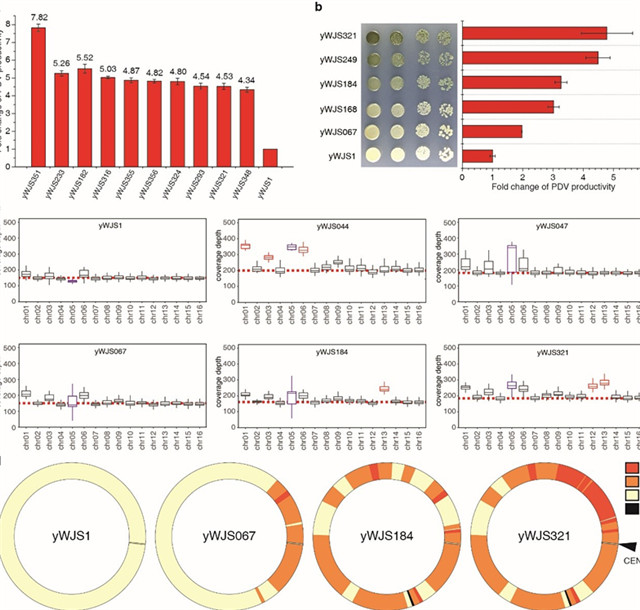
Figure. 2 Continuous improvement of PDV production and generation of structural variations.
“The SCRaMbLE method is generally useful for alteration of defined phenotype and genotype with a ring chromosome genetic background, especially for genome reorganization”, said Professor Ying-Jin Yuan, “Our continuous approach by SCRaMbLEing ring_synV embraces the evolution of the genome by modifying the chromosome number, structure and organization, and evolution of organisms with improved properties, identifying targets for phenotypic comprehension.”
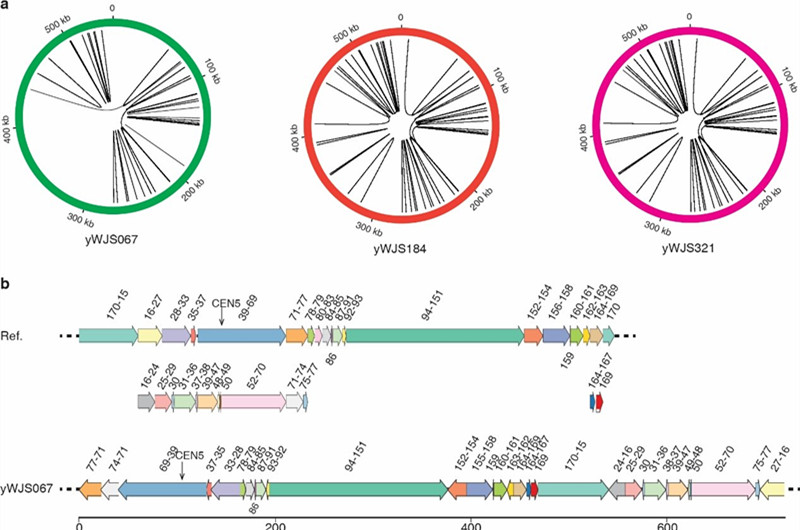
Figure. 3 Intrachromosomal interactions in the SCRaMbLEd ring_synV.
By: Mian Qin
Editor: Keith Harrington






